
- #PCR TROUBLESHOOTING BAND IN NEGATIVE CONTROL HOW TO#
- #PCR TROUBLESHOOTING BAND IN NEGATIVE CONTROL CODE#
This was different for us since we were used to multiplying it by two for the master mix. To be able to start our temperature gradient, we had to multiply our PCR calculations by nine. We run this process through a thermal cycler, which takes the PCR’s through temperatures that denature, anneal, and extend the DNA in the samples.Īveya and Madisyn pipetting the 163 male brain cDNA into the positive control tubes. We thought that this would reduce our primer dimers and give us clearer results. We ran the temperature gradient on our third primer set only because it was the only primer set that gave us decent results throughout the troubleshooting process. Cohen about our results, we were instructed to run a temperature gradient to see if there are more optimal annealing temperatures. Our first two primer sets were amplifying off large targets. We did not see that in the majority of the PCRs that we ran except for our third primer set.
For our primers, our band sizes should range between 126-154b. Our image revealed primer dimers, incorrect band sizes, and faint bands.
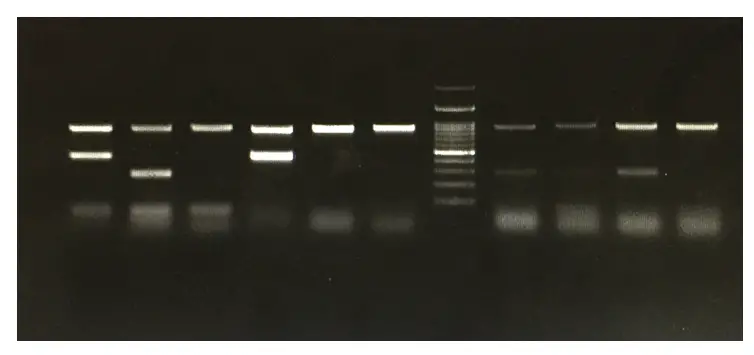
After running our gel and photographing it using a gel imaging bio-doc machine, our results had a lot of issues. For this specific PCR, we used 163 male brain (lizard tissue) as our cDNA. Beta-actin is used as a reliable reference gene. Over the past couple of weeks, the first PCR that we ran contained the three primer sets that we picked and beta-actin, which consisted of a negative control (H2O) and a positive control (cDNA). We did that during our downtime to make sure that we used our time in the lab efficiently. It takes 30 minutes to solidify so with our downtime, we create new PCR master mixes to run the following week. The comb creates the wells that we will pipet our PCR into. Once the liquid is cooled, we can then place it into the gel casting tray with a comb. We then microwave this for around 30 seconds to fully dissolve the agarose. The gel electrophoresis can be made by weighing out 0.75 grams of agarose, which is a powder, and adding 50 ml of 1x TAE to make it a liquid. We stain this gel with a DNA-binding dye, and the DNA fragments will be shown as bands. We can observe our primers through gel electrophoresis, which is a technique that will separate DNA fragments by their size. PCR is used to amplify specific DNA sequences. Gus pipetting water (our negative control in this experiment) into the master mix solutions. For the upcoming weeks we will perform gel extraction and continue our research.
#PCR TROUBLESHOOTING BAND IN NEGATIVE CONTROL HOW TO#
Throughout this process, we learned how to run PCR, agarose gel, PCR-clean up, maintain a lab notebook, expand our knowledge, and improve our skills. At the beginning of our troubleshooting, we had primers dimers, so some of our results were not as great as we expected. Our lab work is going good so far and we still have some things to do. The first well is our ladder, the second well is our positive control, and the third well is our negative control. After we get our results, we will analyze the sequencing results. After we finished the PCR cleanup, we placed our samples in the freezer and Dr. We also did calculations to figure out how much water we will add to the PCR that we will send for sequencing. These numbers show how much pure DNA we had in our solution. We used nanodrop to record the DNA concentration and the purity ratio from our solution. We cut the band from the gel and placed it in a microcentrifuge tube, melted the gel, and then took several steps to wash away any undesired material from the DNA.
#PCR TROUBLESHOOTING BAND IN NEGATIVE CONTROL CODE#
If the sequence matches the code for the HS6ST1 gene, we know that primer set two is amplifying the desired DNA sequence. The goal of the PCR clean-up was to create a solution with pure DNA from our gel that can be sequenced. The next step was to move forward with a gel extraction to perform PCR-cleanup after spring break.
The results for the PCR showed that there still were primer dimers in our second set, but there was a nice band to work with. There were some primer dimers on our gel, so we modified the concentration of our primer set from 1.0µM to 0.5µM and ran PCR and agarose gel for primer set two. We found out that our second primer set was giving us a nice band on the gel, so we moved forward with that set. Ilhan loading a primer set 2 PCR into a gel.


 0 kommentar(er)
0 kommentar(er)
